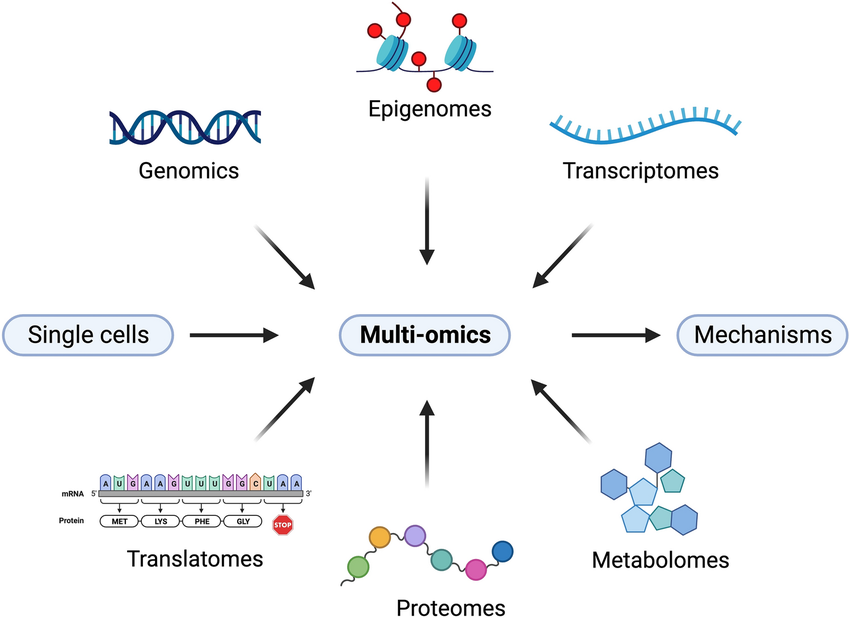
Single-cell and spatial multi-omics technologies allow researchers to study individual cells in their spatial context. These approaches enable the detection of molecular heterogeneity, cell state dynamics, and tissue organization with high resolution. This document presents a comprehensive technical overview of key methods, current applications, and emerging directions.
Definition and Scope Single-cell omics refers to profiling genomic, transcriptomic, proteomic, and epigenomic features at the single-cell level. Spatial omics integrates molecular profiling with positional information of cells within intact tissues.
Reference: Introduction to Omics Technologies - NCBI Reference: Single-Cell Analysis Overview - NIBIB
Key Single-Cell Omics Methods
2.1 Single-Cell RNA Sequencing (scRNA-seq) Enables quantification of gene expression in individual cells. Platforms: 10x Genomics Chromium, Drop-seq, Smart-seq2. Challenges: dropout events, technical noise, batch effects.
Reference: scRNA-seq Guide - Broad Institute
2.2 Single-Cell ATAC-seq Assesses chromatin accessibility, identifying active regulatory elements. Useful for studying gene regulation and cell identity.
Reference: scATAC-seq Overview - NCBI Reference: ATAC-seq Protocols - UCSD
2.3 CITE-seq and REAP-seq Combine RNA sequencing with protein detection using DNA-barcoded antibodies.
Reference: CITE-seq Method - Rockefeller University Reference: REAP-seq Overview - NCBI
2.4 Joint Multi-Omics Profiling Methods: SNARE-seq (RNA + chromatin), scTrio-seq (RNA + DNA methylation + CNV).
Reference: scMulti-omics Techniques - Nature Biotechnology
Key Spatial Omics Methods
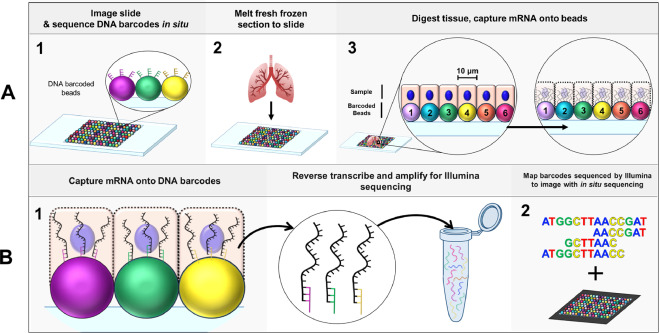
3.1 Spatial Transcriptomics Detects spatial gene expression patterns in tissue sections. Technologies: 10x Visium, MERFISH, seqFISH, Slide-seq.
Reference: Spatial Omics at Human Cell Atlas
3.2 Imaging Mass Cytometry (IMC) Uses metal-conjugated antibodies and laser ablation to visualize protein distribution.
Reference: IMC Overview - NCBI Reference: CyTOF Resource - Stanford University
3.3 CODEX Imaging Cyclic immunofluorescence imaging for multiplex protein detection.
Reference: CODEX Imaging - Akoya Biosciences
3.4 Spatial Metabolomics Mass spectrometry-based imaging (e.g., MALDI-IMS) for mapping metabolites.
Reference: Metabolomics Program - NIH Reference: MALDI Imaging Overview - NCBI
Applications
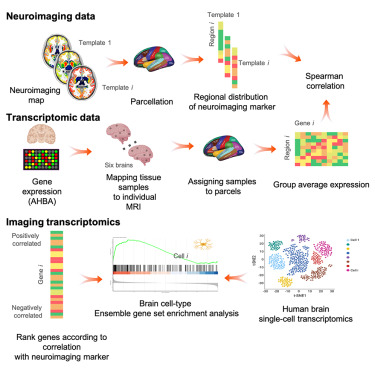
4.1 Neuroscience Mapping transcriptomic and epigenomic profiles of neuronal subtypes. Integration with connectomic data.
Reference: NIH BRAIN Initiative Reference: Allen Brain Atlas
4.2 Cancer Research Tumor heterogeneity analysis. Characterization of tumor-immune interactions.
Reference: Omics Tools in Cancer - National Cancer Institute
4.3 Immunology Profiling immune cell states across disease and vaccination conditions.
Reference: Human Immunology Project - NIAID Reference: Immune Gene Data - NCBI
4.4 Developmental Biology Lineage tracing and spatial mapping in embryos.
Reference: EBI Development Atlas Reference: Xenopus Development Data - NCBI
4.5 Regenerative Medicine Identifying regenerative states and guiding tissue repair.
Reference: NIH Regenerative Medicine Program
Emerging Concepts
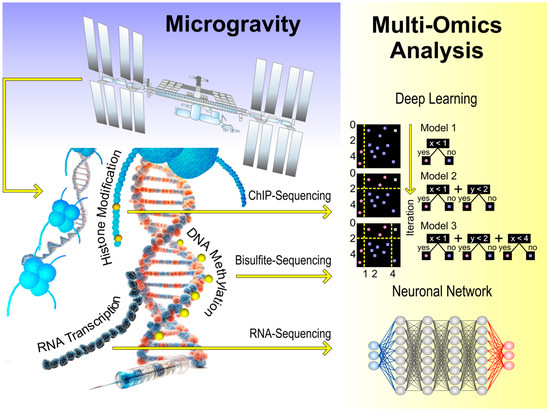
5.1 Space Biology Applying omics technologies to understand biological responses in microgravity.
Reference: NASA GeneLab
5.2 AI-Driven Omics Design Use of machine learning to optimize experimental design and omic layer selection.
Reference: NIH Bridge2AI Program Reference: Bioinformatics Tools - NCBI
5.3 Live-Cell Omics Development of non-destructive techniques for dynamic molecular profiling.
Reference: Live-Cell Imaging Research - NIBIB
5.4 Digital Tissue Twins Building 3D digital reconstructions of tissues integrating multi-modal data.
Reference: HuBMAP Consortium
5.5 Environmental Applications Applying spatial omics in soil, ocean, and microbiome contexts.
Reference: Human Microbiome Project - NIH Reference: DOE Joint Genome Institute
Single-cell and spatial multi-omics provide detailed insight into cellular heterogeneity and tissue structure. These technologies are critical in modern biology and will shape future biomedical research and precision medicine.
🔬 Event Spotlight: SCSOMICS 2025 – The Single-Cell Revolution Continues
The field of single-cell and spatial multi-omics is advancing rapidly, and one of the key gatherings for scientific exchange is the upcoming event:
SCSOMICS 2025: The Single-Cell Revolution Continues
📍 Date: 2025
📍 Location: Online and in-person hybrid
📍 Focus: Advanced single-cell technologies, data integration, spatial resolution, and clinical applications
This event brings together leading scientists, developers, and clinicians to explore innovations in multi-omics analysis, including AI-driven interpretation, high-throughput platforms, and translational case studies.
Topics include:
- Spatial transcriptomics and multiplex imaging
- Integrative single-cell omics workflows
- Clinical and pharmaceutical case applications
- Emerging automation in cell profiling
Researchers interested in the practical implementation of cutting-edge omics technologies and future collaborations are encouraged to register here.