High-Density Genomic Analysis with BeadArray Technology
The BeadArray technology is a powerful method used for high-density genomic analysis. It utilizes three-micron beads coated with DNA capture probes that are randomly self-assembled into etched wells on a substrate. The technology is deployed on platforms consisting of fiber optic bundles, each containing numerous individual fiber strands. These fiber bundles are etched to form microscopic wells, each holding a single bead coated with DNA probes.
"Beads in Wells" SEM Showing 3-Micron Beads in Wells
This technology is central to high-density array platforms, with its application across different array configurations. The 3-micron beads are arranged in etched wells, enabling highly efficient genomic analysis.
The process begins by adding a liquid suspension of beads to the etched fiber face of the bundle. Each well randomly captures one bead, which ensures a high-density array where each bead type is represented multiple times. The assembled array is then exposed to a fluorescently labeled cDNA sample. The sample strands bind to their complementary probe DNA, while strands without a match do not bind.
This technology allows for efficient multi-sample analysis in a format known as the Array of Arrays. The configuration includes multiple fibre bundles, enabling the interrogation of hundreds to thousands of genetic targets per sample, generating large datasets for each experiment. The throughput of this system is considerably higher than many other platforms, making it suitable for large-scale genomic studies and applications.
HapMap Project
The International HapMap Project was launched with the aim of accelerating the discovery of genes linked to common diseases such as asthma, cancer, diabetes, and heart disease. The goal of this global initiative was to create a comprehensive map of genetic variation. As part of this project, one organization participated with a focus on advancing the development of genomic analysis technologies.
At the same time, it became clear that the existing collaboration was not meeting expectations due to limitations in multiplexing capabilities. The initial assay technology could not achieve the high levels of multiplexing that the new platform could support.
In response, the organization had already developed a powerful assay capable of multiplexing at higher levels, as well as a high-resolution BeadArray Reader for imaging small features. They also developed a high-throughput genotyping system and associated laboratory information management protocols. These protocols were submitted for the project’s grant application, which led to a significant role in the HapMap initiative.
The technology contributed to the genotyping of a substantial portion of the HapMap dataset, representing a significant contribution to the global effort. Moreover, most major investigative groups involved in the project utilized the developed system, making the technology a key component in the success of the initiative.
A New BeadArray Platform
The second microarray platform introduced was a new BeadChip technology. This platform provided researchers with greater flexibility, allowing for projects to be conducted on a smaller sample scale or at higher density, such as whole-genome analyses, without the need for production-scale facilities.
The platform supports multiple configurations of BeadChips, including options that can analyze over 46,000 or 23,000 transcripts per sample, with the ability to handle multiple samples simultaneously. Some configurations feature over 10 million features per microarray.
These innovations were the first of their kind in the industry, enabling whole-genome expression analysis across multiple samples on the same microarray. The technology offered high sensitivity, selectivity, and reproducibility, as well as the advantage of low reagent volumes and cost-effective per-sample pricing. The introduction of these BeadChips expanded their applicability, with further developments planned for additional species, such as mouse and rat.

The DASL Assay
The technology has recently been applied to an assay known as DASL™ (Annealing, Selection, Extension, and Ligation), a key feature of which is its ability to enable RNA from formalin-fixed, paraffin-embedded (FFPE) tissue samples to be used effectively.
Microarray analysis of gene expression in clinical samples linked to treatment responses and patient outcomes holds the potential to uncover important biomarkers associated with complex diseases such as cancer. However, the availability of high-quality, well-preserved samples has been a significant challenge.
FFPE tissue samples are particularly valuable because they are connected to clinical outcomes and are widely available, with an estimated 400 million FFPE samples archived in North America for cancer research alone. Despite their potential, the degraded RNA in these samples has limited their full utilization. Although degraded RNA samples can be assayed, traditional approaches, such as low-multiplex qPCR, have been expensive and less effective.
Traditional RNA array methods rely on in vitro transcription, where RNA is primed off the poly (A) tail from the 3' end of the transcript, followed by reverse transcription. However, this process requires the RNA to be intact, as the enzyme must travel along the entire strand. Degraded RNA, which has breaks in the sequence, does not provide a continuous strand for priming, making the use of traditional protocols infeasible.
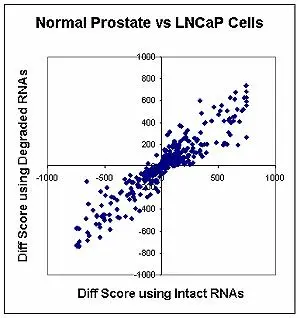
The DASL Cancer Panel was used to measure genes that were differentially expressed between normal prostate tissue and a prostate cancer cell line. Differential expression was compared in samples containing intact RNA and samples with RNA that had been deliberately degraded through heat treatment. The results showed that the set of differentially expressed genes was the same in both degraded and intact samples.
The DASL assay offers an alternative approach to sample labeling. It employs random priming during the cDNA synthesis step, meaning it does not rely on the poly (A) tail. Random 9-base oligos are used as primers, which bind randomly to their sequence throughout the transcription process, enabling reverse transcription to convert mRNA into cDNA.
The key advantage of the DASL assay is that it allows for the efficient analysis of degraded RNA samples. There are large quantities of tissue samples stored in archives, and being able to extract reproducible expression profiles from these degraded samples makes the DASL assay a valuable tool for research.
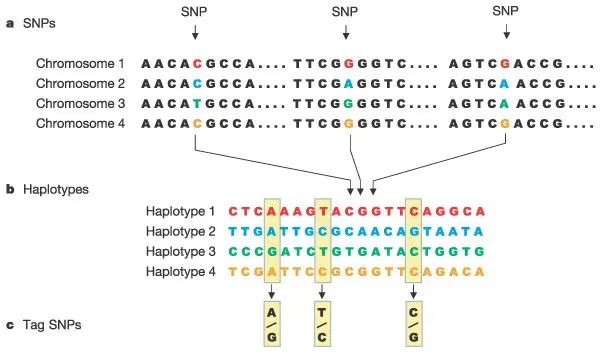
A new assay was introduced for whole-genome genotyping, offering a powerful approach that allows virtually unconstrained locus selection at single-base resolution without the need for PCR. The first microarray to use this assay can query over 100,000 SNP loci, with approximately 70% of these loci located in or near genes, making it particularly valuable for direct disease association studies.
The Infinium Assay
The Infinium assay utilizes the full complexity of the genome, scaling to the number of bead types that can be placed on an array. The assay consists of four main steps: 1) whole-genome amplification, 2) fragmentation and hybridization, 3) enzymatic SNP discrimination by allele-specific extension, and 4) fluorescence staining and scanning.
This powerful approach for whole-genome genotyping allows virtually unconstrained locus selection at single-base resolution, without the need for PCR. The assay is capable of querying over 100,000 SNP loci, with 70% of these loci located in or near genes, offering significant value for direct disease association studies.
Additionally, development efforts are underway for a follow-up product capable of genotyping 250,000 TagSNPs derived from global genetic projects. There are also plans for the release of a one-million-SNP array, along with continued development of specialized assay panels for focused genotyping, such as those targeting gene-rich regions like the MHC (Major Histocompatibility Complex).

The BeadStation Each BeadStation system delivers an end-to-end solution for genetic analysis and constitutes a compelling way to experience the benefits of BeadArray technology.